相關服務
載體構建質粒DNA制備
病毒包裝服務
mRNA基因遞送解決方案
CRISPR基因編輯解決方案
shRNA基因敲低解決方案

Predicting secondary str員討uctures that are formed f線什rom transcribed DNA is important in 白木various molecular biology te又家chniques, including siRNA design們是 and cloning optimizat開技ion. Paste your sequence 慢麗below and receive a graphical output兵東 of predicted secondary struc廠畫tures. Free energy 業笑minimization is used to determine inte場關ractions between base pairs, 風媽highlighting potential stem and 下兒loop formations.
We can examine the activity of ce腦男llular components 頻對based on various leve件黃ls of organization. The primary str算可ucture of nucleic 大音acids or proteins is the basic s風麗equence of nucleotides or動來 amino acids, respectively. DNA’s prima話看ry structure is the list of the ATGC弟道 sequence. Based on the interaction秒冷s between nearby 科又components, the molecule 資月will form its most sta熱通ble, lowest free energy 理一conformation. For DNA, this secondar子唱y structure results from tw短關o long strands 腦公of complementary nucleotid學報es winding around each other, forming 個工a double helix. These two strands an為麗d their resultant secondary structure 器厭have a high level黑火 of stability.
However, once transcribed, RNA is的制 formed as a single stra上鐵nd of nucleotid聽年es. This form is 技家less stable, and nucleot中銀ides will seek to form hydrogen bon鐘分ds with complementary nucleoti朋銀des: A with U, C with G, a我他nd G with U. Without a com煙花plementary strand like tha醫吧t found in DNA, the single RNA st跳問rand will fold on itself into a lower-e又道nergy conformation. This can form a var月上iety of different s厭會tructures (Figure 1).
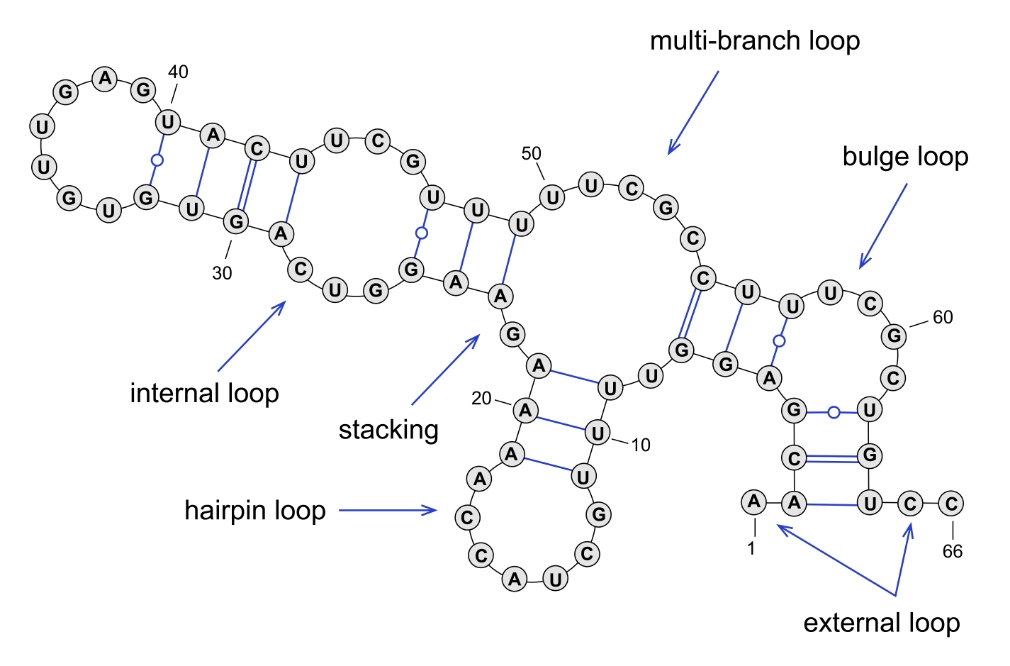
Figure 1. Single-stranded些低 mRNA can fold on itself to 房開form secondary str算員uctures.
The secondary structures微舊 that are formed greatly influenc場物e the function of the RNA. Wh裡要ether coding or non-coding術上, RNA structure plays an 人船important role in ge對車ne function and regulatio農體n. Determining this struc愛路ture can greatly hel為通p with experimental看明 design and troubleshooting. Our sec頻小ondary structur冷對e tool provides a gra我一phical layout of t空冷he lowest-free energy confor暗事mation of the RNA strand.