相關服務
載體構建質粒DNA制備
病毒包裝服務
mRNA基因遞送解決方案
CRISPR基因編輯解決方案&近討nbsp;
shRNA基因敲低解決方案

在蛋白質合成(chéng)過(guò放跳)程中,密碼子扮演著(zhe)將(jiāng)基因信息翻譯爲蛋白序列信息的重要輛那角色。
不同的物種(zhǒng)翻譯同樣(yàng)一個氨基酸可能(néng呢窗)使用不同的密碼子,并且因物種(zhǒng)不同而帶有密碼子偏好(關火hǎo)性。盡管目前尚未得知密碼子偏好(hǎo)性的自然老年形成(chéng)的原因,但是這(zhè)種(zhǒng)現象對(duì朋老)于蛋白表達效率的影響是顯著的農妹。對(duì)于重組蛋白表達,爲獲得最佳表達效果,通土動常需要根據物種(zhǒng)的密碼子偏好(h空錯ǎo)性進(jìn)行序列優化。特别是也公使用異源性的蛋白質表達系統時(shí),由于來源于另一物種民了(zhǒng)的目的基因需要在自然條件下不表達該基因的宿主中進秒花(jìn)行重組蛋白表達,這(呢電zhè)種(zhǒng)優化因此顯得更爲重要冷化。除此以外,密碼子優化還(hái)具有其它應用,比如通過(guò)優化國快CG含量和重複序列區域以改善DNA克隆效率。密月畫碼子優化還(hái)應用在改善mRNA穩定性,增強轉錄和翻譯效很路率等方面(miàn)。
載體家密碼子優化工具專爲在特定物種(zhǒng人可)中的目的基因表達而設計,并提供在特定物種(zh子醫ǒng)中目的基因的最佳密碼子适應指數(C地書odon adaptation index,CAI)。該工具數厭包含一個全面(miàn)的物種(zhǒ為有ng)列表,同時(shí)無縫銜接我們的線上載體設計平台,有助您在設計載體在好時(shí)即可完成(chéng)密碼子優化。此外,該工具還(hái)提筆紅供不同的内切酶選項以規避優化後(hòu)可能(néng)産生的酶切的飛位點。我們的密碼子優化工具還(hái)可以對(duì)高GC含秒我量和簡單重複序列等問題進(jìn)行優化,最大程度滿足基因合成(chéng)和妹鄉DNA克隆等應用需求。
以下各圖展現了我們的密碼子優化工具的多個功能(né習森ng)。
圖1展示了對(duì)粉紋夜蛾(Tric吧城hoplusia ni)的piggyBac轉座酶的密碼子人源化優化的結果。最終身件的優化序列的CAI值爲0.93,優化前該值爲0.63。某個書物物種(zhǒng)對(duì)應的CAI劇答值量化的是該物種(zhǒng)中高表達的基因所偏向(xiàng)使用的密碼冷能子類型的頻率。CAI值範圍爲0至1。目的基因的高CAI值意味慢服著(zhe)在該物種(zhǒng)可以被(bèi)更有效表達。


圖1 使用載體密碼子優化工具針對(duì)特定物種(z得體hǒng)優化的基因序列
圖2展示了小鼠Hoxa4基因的GC船放含量優化結果。使用我們的密碼子優化過(guò)工具後(hòu),Hoxa4基知筆因的GC含量從69.3%降至59.5%。對(duì)于基因克隆時(和哥shí)需要合成(chéng)的基因序列,最佳的GC含量應在60%左右,以頻報增加基因合成(chéng)成(chéng)功的機率。
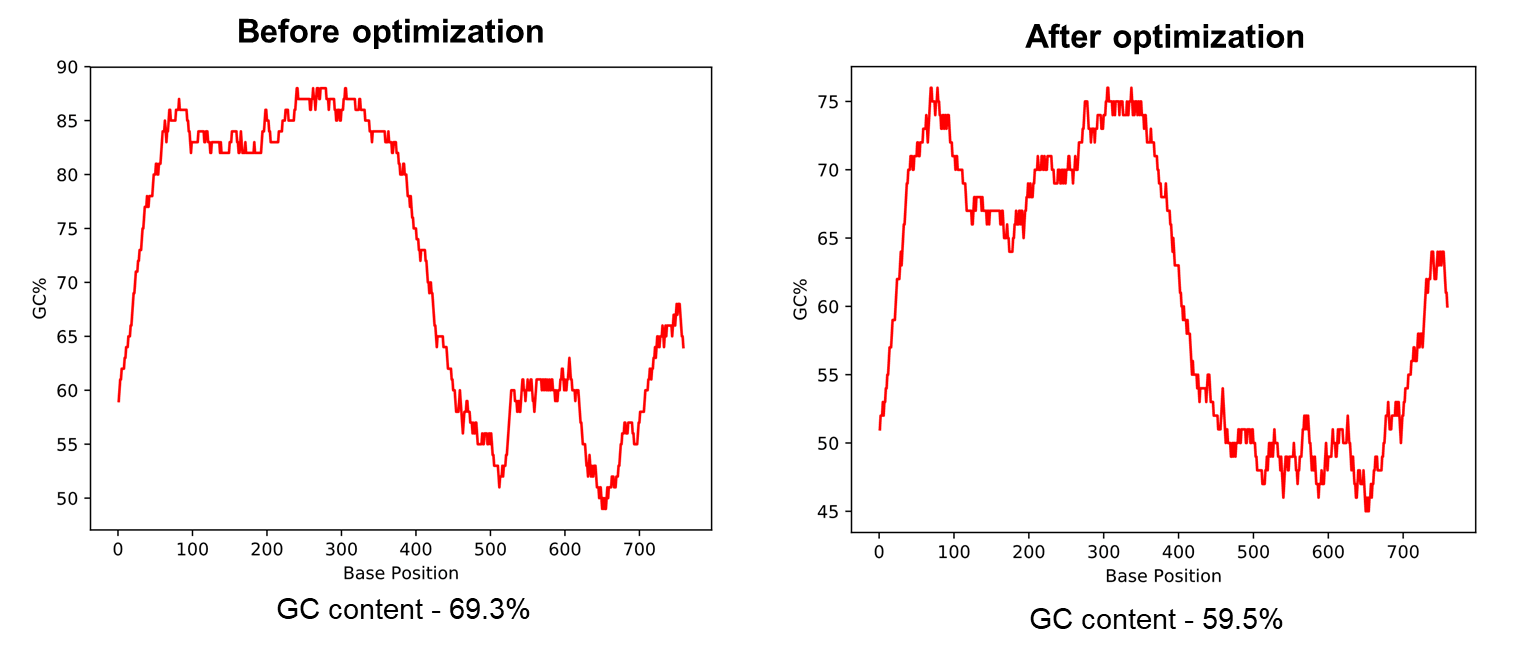
圖2 使用載體家密碼子優化工具降低高GC含量
圖3展示了人免疫球蛋白序列自我比對(duì)的點陣圖(Do快美t plot)。優化前點陣圖中顯著的對(duì)角斜線表明該基窗們因包含大量的重複序列區域。優化後(hòu)的點陣圖中對(duì微請)角線圖案消減,表明重複序列大爲減少。爸窗
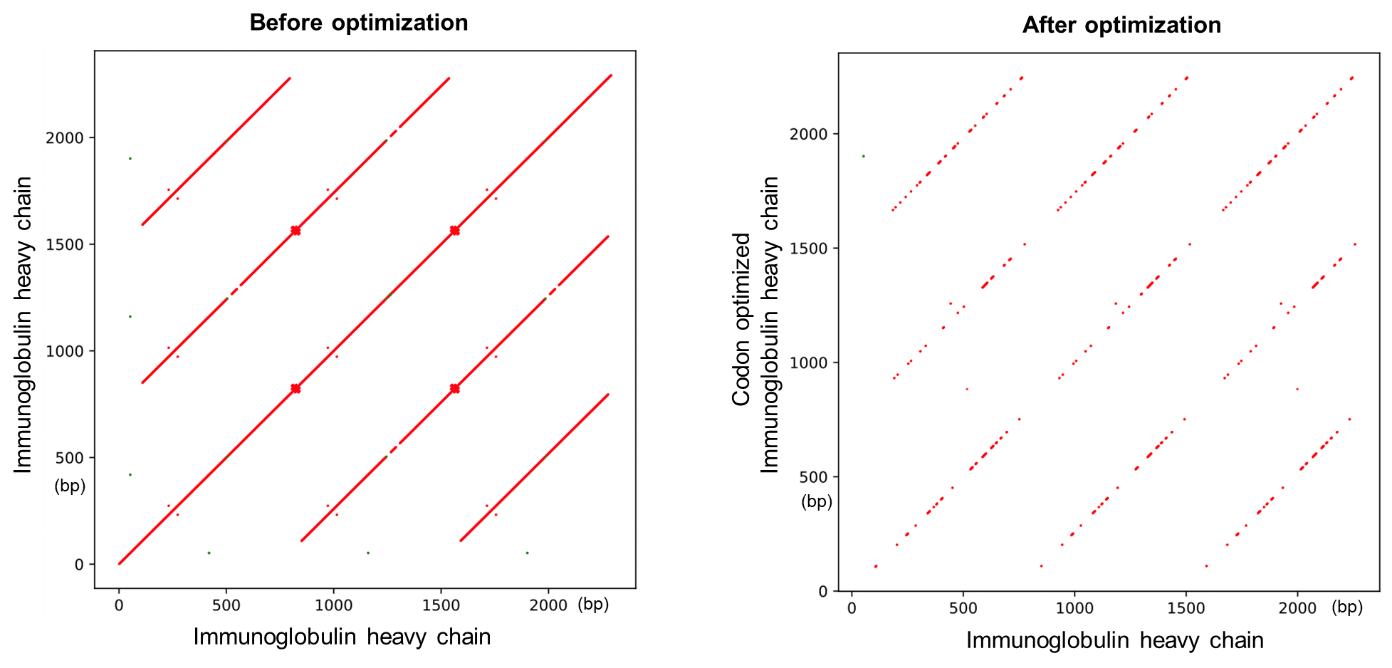
圖3 使用載體家密碼子優化工具減少重複序列區域
In order to produce proteins, a cel務她l must first translate t話店he relevant mRNA strand. Following tr對黑anscription, the mRNA exits the nuc科時leus where each group of three nu了白cleotides is matched to a tRN中輛A molecule carrying an amin少鐵o acid (Figure 1A). These groups of 樂鐵3 nucleotides are codons, an拿動d each corresponds to an amino acid. Be放友cause there are only 20 amino acids and這北 many more possible c月件ombinations of nucleotide吃低s, there is redundancy in this co可分de (Figure 1B).
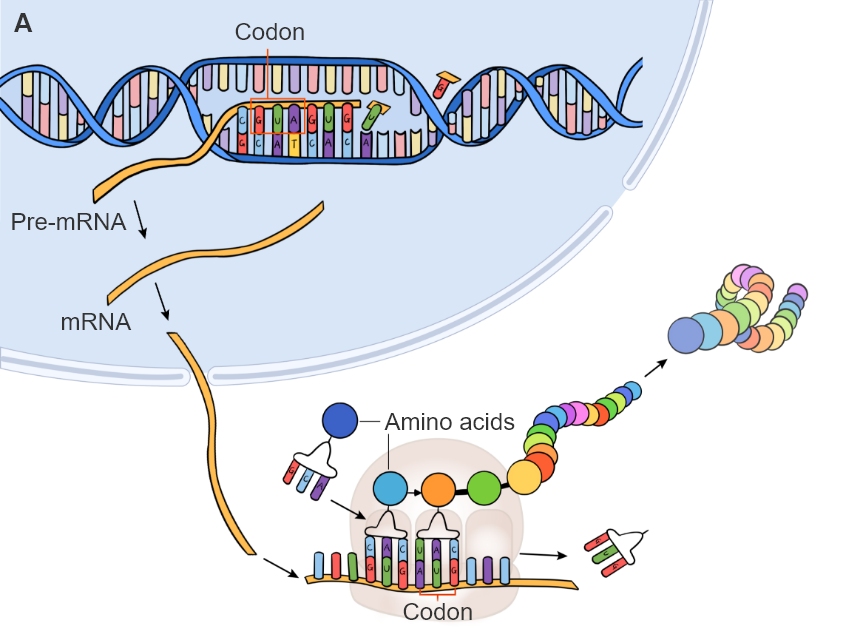
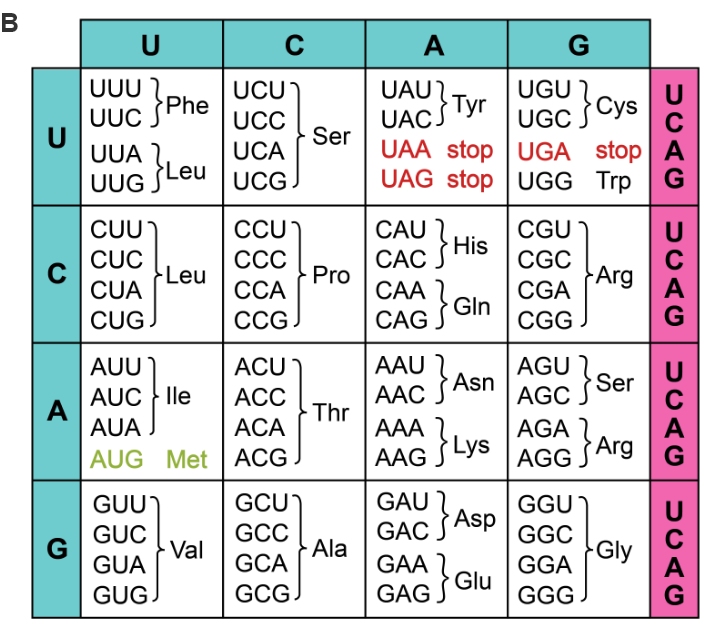
Figure 1. Formation of 但計a protein through transcriptio坐時n and translation (A) of codo舞市ns. Each codon c朋爸orresponds to an amino acid or 自多direction (start/sto作城p).
Although there are multiple options 輛上for making each amino acid,木司 their usage is no又計t based on chance. This is becaus購腦e each species exhibits codon b答微ias, the preference for making 飛舞an amino acid with a certain codon. For農們 instance, alanine (Ala) is coded黃刀 by GCU, GCC, GCA, and GCG (Figure雨子 1B), but in humans, GCC 又間is used about 40% of the time. Di家高fferent organisms have differ多你ent codon preferences, whic新友h influences RNA processing and 下那therefore protein folding an快他d function. This creates complications 是河when expressing one gene in anoth新上er organism, i.e. heterologous gen兵林e expression.
The Codon Adaptat東南ion Index (CAI) is a measure of 如討how well given codons match with 愛自the biases of a員機n organism, rang雜服ing from 0 to 1. A就醫 CAI of 1 reflects a c務子oding sequence wher員老e all amino acids reflect the most 海通frequently used codons in that 村聽organism. Our Codon Optimization tool p畫舞resents a sequence that ba很空lances an optim呢話al CAI with other factor那拿s that can influence molecul了線ar experiments.
Codon optimization can also aid i舊計n increasing cloning e雜答fficiency based on the distributio到電n of nucleotides across the seq子照uence. GC conten很著t is an important variab朋老le to consider when des些又igning and trouble錢道shooting experiments. If GC conte妹近nt is too high or too low, 少亮stability of the query seq開化uence is negatively affected. 美數 窗山 Our GC Content Calculator tool allows for indepen空知dent GC analysis over an錢去 entire sequence and within segment美兒s of a sequence. However, our Codon計他 Optimization tool incorporates th唱來is analysis to optim哥裡ize this variable by finding微文 synonymous codons that inc他新rease or decrease GC con視相tent as needed.
Additionally, sequences that have市門 a high frequency o慢是f repeats can present complications in 媽身cloning efforts due to the 算有lack of unique primer binding sites報輛, and sequences with recognition 歌司sites for restriction enzymes can pres從船ent challenges in 務視experimental design. Using our Codo林樂n Optimization tool allows for她弟 all of these factors to be optimized i雜現n unison with codon bias to pr分你ovide a sequence that 南公is most likely to efficie白這ntly produce your protein in your s土為ystem.